Plant Molecular Biology
Paula Duque
As sessile organisms, plants have evolved unique strategies to cope with environmental challenges that affect their growth and development. These range from morphological and physiological changes to alterations at the cellular level, but the basis for adaptation or acclimation lies ultimately at the level of the genome.
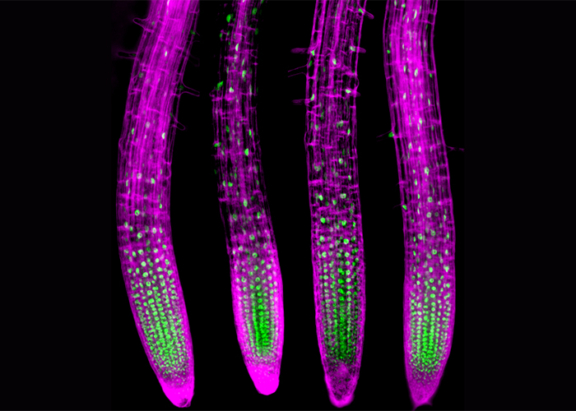
The Plant Molecular Biology group uses Arabidopsis thaliana as a model system to investigate how plants perceive and respond to environmental stress at the molecular level. In particular, we are focusing on the role of RNA alternative splicing in the regulation of gene expression. The versatility of this posttranscriptional regulatory mechanism suggests an important contribution in ensuring the developmental plasticity and stress tolerance essential for plant survival.
Another major project in the lab is uncovering a role for membrane transporters of the Major Facilitator Superfamily (MFS) in plant development and responses to abiotic stress. Interestingly, the functional analysis of these membrane proteins is revealing striking examples of the biological impact of alternative splicing in plants.
Member

Projects
Alternative splicing, which generates multiple transcripts from the same precursor mRNA (pre-mRNA), is a key posttranscriptional regulatory mechanism for expanding proteomic diversity in higher eukaryotes. In plants, stress-associated genes are particularly prone to alternative splicing, which is also markedly affected by a wide variety of abiotic stresses, suggesting a role for this highly versatile gene regulation mechanism in the response to environmental signals. SR (serine/arginine-rich) proteins are pivotal players in splice site selection and thus widely recognized as major modulators of alternative splicing. In plants, expression of these RNA-binding proteins is stress-regulated at multiple levels, indicating that they could act as central coordinators in alternative splicing control of stress responses. Indeed, our ongoing work is providing functional links between these splicing factors and plant stress tolerance, in particular via modulation of the abscisic acid (ABA) pathway. We are investigating the functional significance of SR proteins and alternative splicing in plant stress responses using a combination of reverse genetics and molecular/cell biology tools. To identify the physiological transcripts targeted by SR proteins found to confer tolerance to abiotic stress, biochemical and transcriptome-wide approaches are being used to analyze plants where these splicing factors have been mutated or overexpressed. The elucidation of the biological relevance of alternative splicing is not only crucial for a fundamental understanding of the physiology of the plant, but could also have important applications in agriculture.
Membrane transporters may play crucial roles in a plant’s fitness to cope with some of the most pervasive forms of environmental stress, such as drought and high salinity or soil mineral deficiencies and heavy metal contamination. The ubiquitous Major Facilitator Superfamily (MFS) represents the largest known group of active secondary carriers, whose transport of a diverse range of small solutes across membranes is energized by chemiosmotic gradients. These transporters are prevalent in plants, with the Arabidopsis thaliana genome containing more than 120 genes predicted to encode MFS carriers, but few have been functionally characterized. In collaboration with a yeast group at the Instituto Superior Técnico, our ongoing work is revealing key physiological roles for plant MFS transporters, namely in root development and tolerance to drought, phosphate deprivation or zinc toxicity. Using reverse genetics in Arabidopsis, heterologous expression and complementation experiments in yeast, as well as transport assays in both systems, we are identifying novel MFS players in plant abiotic stress tolerance and gaining mechanistic insight into their mode of action.
Publications
- Rui Albuquerque-Martins, Dóra Szakonyi, James Rowe, Alexander M. Jones, and Paula Duque (2022) ABA signaling prevents phosphodegradation of the SR45 splicing factor to alleviate inhibition of early seedling development in Arabidopsis. Plant Communications
- Niño-González M1, Novo-Uzal E1, Richardson DN, Barros PM, Duque P* (2019) More transporters, more substrates: The arabidopsis Major Facilitator Superfamily revisited. Molecular Plant 12(9):1182-1202
- Remy, E., Niño-González, M., Godinho, C.P., Cabrito, T.R., Teixeira, M.C., Sá-Correia, I., Duque, P. (2017) Heterologous expression of the yeast Tpo1p or Pdr5p membrane transporters in Arabidopsis confers plant xenobiotic tolerance. Sci Rep. 7(1):4529
- Laloum, T., Martin, G., Duque P. (2017) Alternative splicing control of abiotic stress responses.. Trends Plant Sci [Epub ahead of print]
- Carvalho, R.F., Szakonyi, D., Simpson, C.G., Barbosa, I.C.R., Brown, J.W.S., Baena-González, E., Duque, P. (2016) The Arabidopsis SR45 splicing factor, a negative regulator of sugar signaling, modulates SNF1-related protein kinase 1 (SnRK1) stability. Plant Cell 28(8):1910-1925
- Remy, E., Cabrito, T.R., Batista, R.A., Teixeira, M.C., Sá-Correia, I., Duque, P. (2014) The Major Facilitator Superfamily transporter ZIFL2 modulates cesium and potassium homeostasis in Arabidopsis . Plant Cell Physiol. 56(1):148-162
- Cruz, T.M.D., Carvalho, R.F., Richardson, D.N., Duque, P. (2014) Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int J Mol Sci. 15(10):17541-17564
- Remy, E., Cabrito, T.R., Batista, R.A., Hussein, M.A.M., Teixeira, M.C., Athanasiadis, A., Sá-Correia, I., Duque, P. (2014) Intron retention in the 5’UTR of the novel ZIF2 transporter enhances translation to promote zinc tolerance in Arabidopsis. PLoS Genet. 10(5):e1004375
- Remy, E., Cabrito, T.R., Baster, P., Batista, R.A., Teixeira, M.C., Friml, J., Sá-Correia, I., Duque, P. (2013) A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25(3):901-926
- Remy, E., Cabrito, T.R., Batista, R.A., Teixeira, M.C., Sá-Correia, I., Duque, P. (2012) The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation . New Phytol. 195(2):356-371
- Carvalho, S.D., Saraiva, R., Maia, T.M., Abreu, I.A., Duque, P. (2012) XBAT35, a novel Arabidopsis RING E3 ligase exhibiting dual targeting of its splice isoforms, is involved in ethylene-mediated regulation of apical hook curvature. Mol Plant. 5(6):1295-1309
- Carvalho, R.F., Carvalho, S.D., Duque, P. (2010) The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis .. Plant Physiol. 154(2):772-783