Chromosome Dynamics
Raquel Oliveira
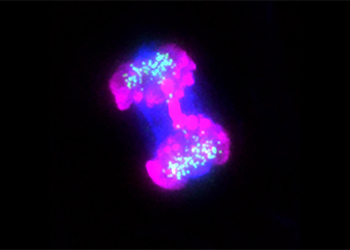
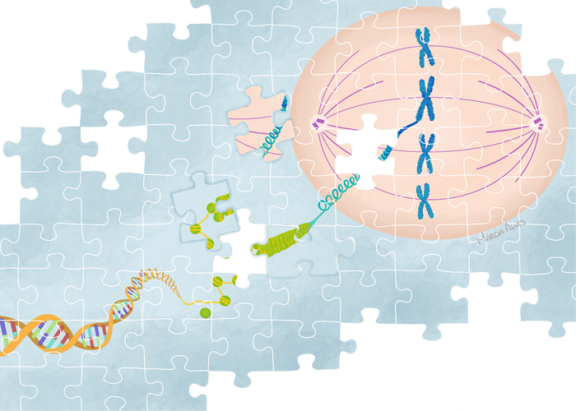
In Raquel Oliveira Lab, researchers study how chromosome architecture contributes to faithful genome segregation. Genome stability relies on the fact that at each round of cell division, the genetic information encoded in the DNA molecules is properly segregated into the two daughter cells. Proper completion of this process, in turn, depends on two major changes in chromosome organization:
- The two-sister DNA molecules remain tightly associated with each other from the moment of DNA replication until the later stages of the subsequent mitosis;
- At the onset of nuclear division, chromatin is converted into compact structures with the right mechanical properties (size, flexibility, and rigidity) to facilitate their segregation.
The laboratory adopts a multidisciplinary approach, combining Drosophila genetics, acute protein inactivation, 4D-live cell imaging and biophysical/mathematical modelling to evaluate how dynamic mitotic chromosomes are assembled and how their morphology influences the mechanical aspects of chromosome movement and cell cycle checkpoint signalling.
In parallel they aim to dissect how different cells respond to compromised chromosome cohesion and condensation, both at the cellular and organism level. By studying the contribution of chromosome structure in the mechanics of nuclear division researchers aim to identify novel routes to aneuploidy that underlie several human conditions, including developmental diseases, cancer and infertility.
Funding

Publications
- Margarida Araújo, Alexandra Tavares, Diana V Vieira, Ivo A Telley and Raquel A Oliveira (2022) Endoplasmic reticulum membranes are continuously required to maintain mitotic spindle size and forces. Life Science Alliance
- Carvalhal S., Ingrid Bader I., Rooimans M.A., Oostra A.B., Balk J.A., Feichtinger R.G., Beichler C., Speicher M.R., van Hagen J.M., Waisfisz Q., van Haelst M., Bruijn M., Tavares A., Mayr J.A., Wolthuis R.M.F., A. Oliveira R.A.,de Lange J. (2022) Biallelic BUB1 mutations cause microcephaly, developmental delay, and variable effects on cohesion and chromosome segregation. Science Advances 8 (3)
- Inês Milagre, Carolina Pereira, Raquel A. Oliveira and Lars E. T. Jansen (2020) Reprogramming of human cells to pluripotency induces CENP-A chromatin depletion. Open Biology 10:200227
- Piskadlo, E., Tavares, A., Oliveira R.A. (2017) Metaphase chromosome structure is dynamically maintained by condensin I-directed DNA (de)catenation.. eLife
- Piskadlo, E., Oliveira R.A. (2016) Novel insights into mitotic chromosome condensation.. F1000Research 5:1807.
- Mirkovic, M., Hutter, L.H., Novák, B., Oliveira, R.A. (2015) Premature sister chromatid separation is poorly detected by the Spindle Assembly Checkpoint due to system-level feedbacks.. Cell Reports 13 (3):469-478.
- Oliveira, R.A., Kotadia, S., Tavares, A., Mirkovic, M., Bowlin, K., Eichinger, C.S., Nasmyth, K., Sullivan, W., Lichten, M. (2014) Centromere-independent accumulation of cohesin at ectopic heterochromatin sites induces chromosome stretching during anaphase.. Plos Biology 12:e1001962.
- Oliveira, R.A., Hamilton, R.S., Pauli, A., Davis, I., Nasmyth, K. (2010) Cleavage of cohesin and Cdk inhibition trigger formation of daughter nuclei.. Nature Cell Biology 12:185 - 192.